6 Dec 2018
Abstract
Type V CRISPR-Cas systems are distinguished by a single RNA-guided RuvC domain-containing effector, Cas12. Although effectors of subtypes V-A (Cas12a) and V-B (Cas12b) have been studied in detail, the distinct domain architectures and diverged RuvC sequences of uncharacterized Cas12 proteins suggest unexplored functional diversity. Here, we identify and characterize Cas12c, g, h, and i. Cas12c, h, and i demonstrate RNA-guided double-stranded (ds) DNA interference activity. Cas12i exhibits markedly different efficiencies of crRNA spacer complementary and non-complementary strand cleavage resulting in predominant dsDNA nicking. Cas12g is an RNA-guided RNase with collateral RNase and single-stranded DNase activities. Our study reveals the functional diversity emerging along different routes of type V CRISPR-Cas evolution and expands the CRISPR toolbox.
[Image]
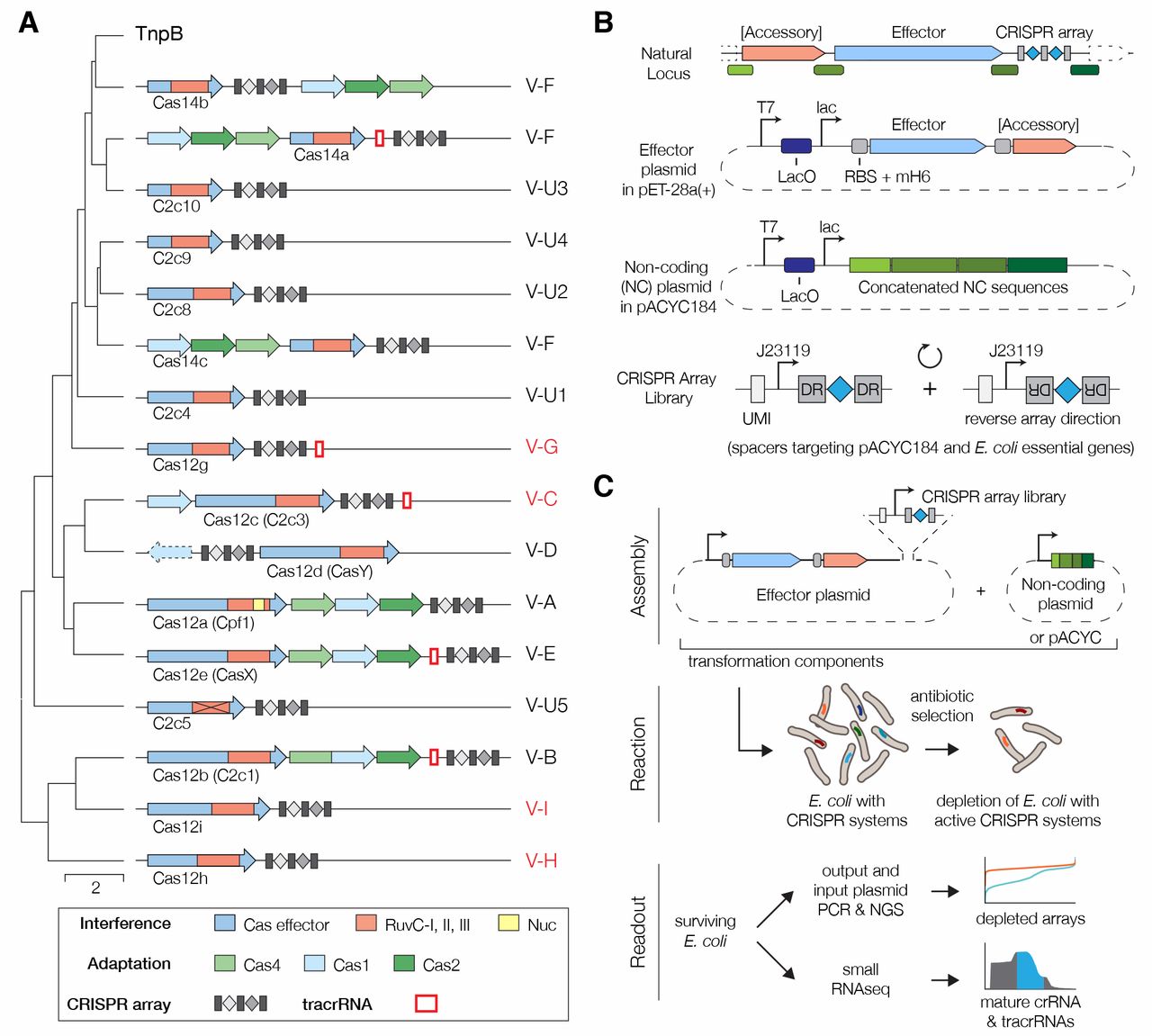
Fig. 1 Discovery and screening of type V CRISPR-Cas diversity.
(A) Classification tree of type V effectors (Cas12 proteins) with corresponding CRISPR-Cas loci organization shown for each branch. Cas12 proteins analyzed in this work are highlighted in red.
(B) Design of in vivo screen effector and non-coding plasmids. CRISPR array libraries were designed with spacers uniquely and uniformly sampled from both strands of pACYC184 or E. coli essential genes, then flanked by two DRs and transcribed by a J23119 promoter.
(C) Workflow schematic of the in vivo E. coli screen.
